I tried looking for a similar question but couldn't find one. If you do please let me know!
I've been working on a project looking at grain staples
Here is a subset of my dataset:
nutrient.component. grain nutrients
1 Beta-carotene (μg) White Rice 0.00
2 Beta-carotene (μg) Brown Rice NA
3 Calcium (mg) White Rice 28.00
4 Calcium (mg) Brown Rice 23.00
5 Carbohydrates (g) White Rice 80.00
6 Carbohydrates (g) Brown Rice 77.00
7 Copper (mg) White Rice 0.22
8 Copper (mg) Brown Rice NA
9 Energy (kJ) White Rice 1528.00
10 Energy (kJ) Brown Rice 1549.00
11 Fat (g) White Rice 0.66
12 Fat (g) Brown Rice 2.92
13 Fiber (g) White Rice 1.30
14 Fiber (g) Brown Rice 3.50
15 Folate Total (B9) (μg) White Rice 8.00
16 Folate Total (B9) (μg) Brown Rice 20.00
17 Iron (mg) White Rice 0.80
18 Iron (mg) Brown Rice 1.47
19 Lutein+zeaxanthin (μg) White Rice 0.00
20 Lutein+zeaxanthin (μg) Brown Rice NA
21 Magnesium (mg) White Rice 25.00
22 Magnesium (mg) Brown Rice 143.00
23 Manganese (mg) White Rice 1.09
24 Manganese (mg) Brown Rice 3.74
25 Monounsaturated fatty acids (g) White Rice 0.21
26 Monounsaturated fatty acids (g) Brown Rice 1.05
27 Niacin (B3) (mg) White Rice 1.60
28 Niacin (B3) (mg) Brown Rice 5.09
29 Pantothenic acid (B5) (mg) White Rice 1.01
30 Pantothenic acid (B5) (mg) Brown Rice 1.49
31 Phosphorus (mg) White Rice 115.00
32 Phosphorus (mg) Brown Rice 333.00
33 Polyunsaturated fatty acids (g) White Rice 0.18
34 Polyunsaturated fatty acids (g) Brown Rice 1.04
35 Potassium (mg) White Rice 115.00
36 Potassium (mg) Brown Rice 223.00
37 Protein (g) White Rice 7.10
38 Protein (g) Brown Rice 7.90
39 Riboflavin (B2)(mg) White Rice 0.05
40 Riboflavin (B2)(mg) Brown Rice 0.09
41 Saturated fatty acids (g) White Rice 0.18
42 Saturated fatty acids (g) Brown Rice 0.58
43 Selenium (μg) White Rice 15.10
44 Selenium (μg) Brown Rice NA
45 Sodium (mg) White Rice 5.00
46 Sodium (mg) Brown Rice 7.00
47 Sugar (g) White Rice 0.12
48 Sugar (g) Brown Rice 0.85
49 Thiamin (B1)(mg) White Rice 0.07
50 Thiamin (B1)(mg) Brown Rice 0.40
51 Vitamin A (IU) White Rice 0.00
52 Vitamin A (IU) Brown Rice 0.00
53 Vitamin B6 (mg) White Rice 0.16
54 Vitamin B6 (mg) Brown Rice 0.51
55 Vitamin C (mg) White Rice 0.00
56 Vitamin C (mg) Brown Rice 0.00
57 Vitamin E, alpha-tocopherol (mg) White Rice 0.11
58 Vitamin E, alpha-tocopherol (mg) Brown Rice 0.59
59 Vitamin K1 (μg) White Rice 0.10
60 Vitamin K1 (μg) Brown Rice 1.90
61 Water (g) White Rice 12.00
62 Water (g) Brown Rice 10.00
63 Zinc (mg) White Rice 1.09
64 Zinc (mg) Brown Rice 2.02
Brown Rice has four NA values.
Based on this graphic,
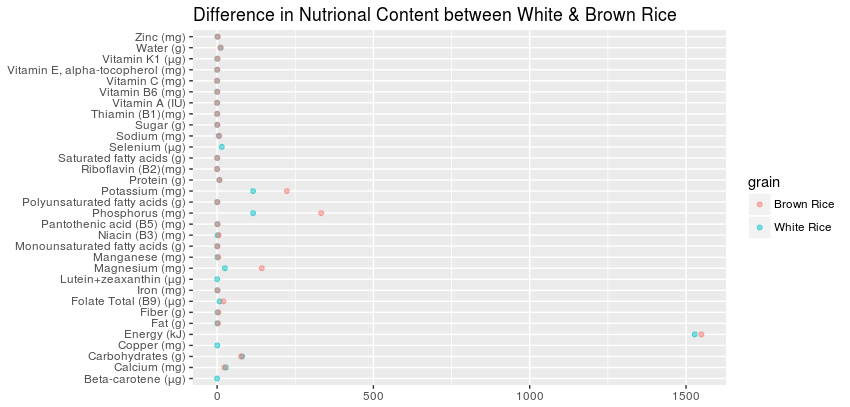 I think it would be fair to assume that the NA values for Brown Rice will be very close to the equivalent value for White Rice. And that it would be more accurate to mirror the white rice values rather than convert the values to zero.
I think it would be fair to assume that the NA values for Brown Rice will be very close to the equivalent value for White Rice. And that it would be more accurate to mirror the white rice values rather than convert the values to zero.
My question is, besides manually looking up and inputting the white rice equivalent nutrient for brown rice, how would the code look to replace NA with the equivalent value for white rice? I would expect the result to convert the NA value for Copper; Brown Rice to be the same value as Copper; White Rice (which is 0.22). Would it be better to first replace NA with zero? But if I do that then I have six nutrients with a value of zero rather than four values with NA. I am trying to figure out the right mindset to solve this through code. Any insight into this would be greatly appreciated.
Thank you

Assuming that the data frame of your input data is called
dt, we can use thefillfunction from thetidyrpackage to achieve this task.dt2is the final output.The default of
fillwill impute theNAbased on the previous and nearest non-NA row. So it is important to make sure that each brown rice record is exactly the next row of the associated white rice record.